Sensors made from custom DNA molecules could be used to personalize cancer treatments and monitor the quality of stem cells, according to an international team of researchers led by scientists at UC Santa Barbara and the University of Rome Tor Vergata.
The new nanosensors can quickly detect a broad class of proteins called transcription factors, which serve as the master control switches of life. The research is described in an article published in Journal of the American Chemical society.
"The fate of our cells is controlled by thousands of different proteins, called transcription factors," said Alexis Vallée-Bélisle, a postdoctoral researcher in UCSB's Department of Chemistry and Biochemistry, who led the study. "The role of these proteins is to read the genome and translate it into instructions for the synthesis of the various molecules that compose and control the cell. Transcription factors act a little bit like the ‘settings' of our cells, just like the settings on our phones or computers. What our sensors do is read those settings."
When scientists take stem cells and turn them into specialized cells, they do so by changing the levels of a few transcription factors, he explained. This process is called cell reprogramming. "Our sensors monitor transcription factor activities, and could be used to make sure that stem cells have been properly reprogrammed," said Vallée-Bélisle. "They could also be used to determine which transcription factors are activated or repressed in a patient's cancer cells, thus enabling physicians to use the right combination of drugs for each patient."
Andrew Bonham, a postdoctoral scholar at UCSB and co-first author of the study, explained that many labs have invented ways to read transcription factors; however, this team's approach is very quick and convenient. "In most labs, researchers spend hours extracting the proteins from cells before analyzing them," said Bonham. "With the new sensors, we just mash the cells up, put the sensors in, and measure the level of fluorescence of the sample."
This international research effort –– organized by senior authors Kevin Plaxco, professor in UCSB's Department of Chemistry and Biochemistry, and Francesco Ricci, professor at the University of Rome, Tor Vergata –– started when Ricci realized that all of the information necessary to detect transcription factor activities is already encrypted in the human genome, and could be used to build sensors. "Upon activation, these thousands of different transcription factors bind to their own specific target DNA sequence," said Ricci. "We use these sequences as a starting point to build our new nanosensors."
The key breakthrough underlying this new technology came from studies of the natural biosensors inside cells. "All creatures, from bacteria to humans, monitor their environments using ‘biomolecular switches' –– shape-changing molecules made from RNA or proteins," said Plaxco. "For example, in our sinuses, there are millions of receptor proteins that detect different odor molecules by switching from an ‘off state' to an ‘on state.' The beauty of these switches is that they are small enough to operate inside a cell, and specific enough to work in the very complex environments found there."
Inspired by the efficiency of these natural nanosensors, the research group teamed with Norbert Reich, also a professor in UCSB's Department of Chemistry and Biochemistry, to build synthetic switching nanosensors using DNA, rather than proteins or RNA.
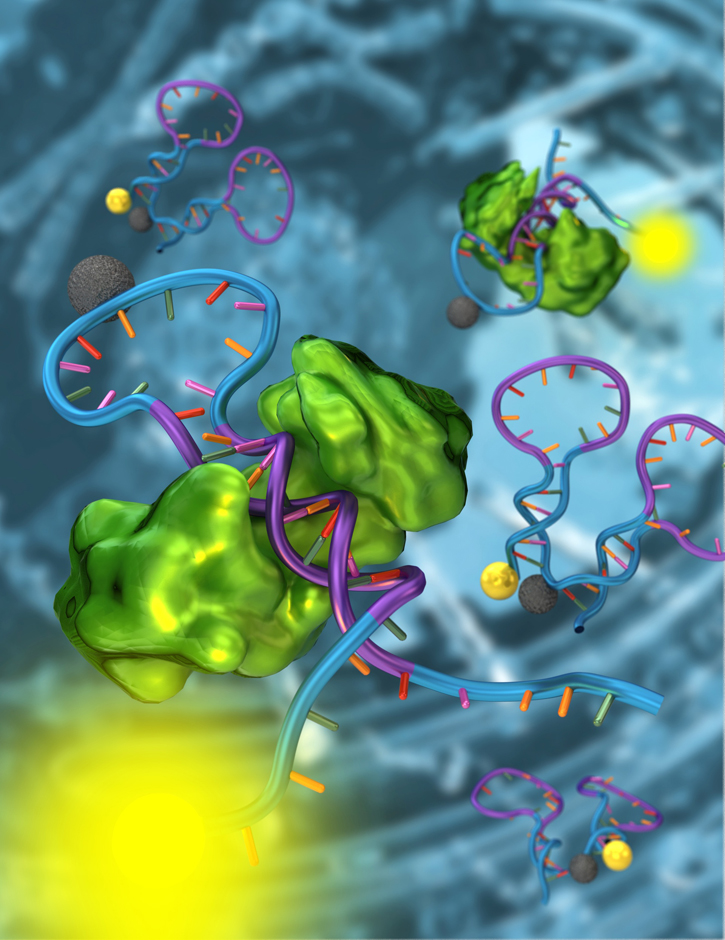
Specifically, the team re-engineered three naturally occurring DNA sequences, each recognizing a different transcription factor, into molecular switches that become fluorescent when they bind to their intended targets. Using these nanometer-scale sensors, the researchers could determine transcription factor activity directly in cellular extracts by simply measuring their fluorescence level.
The researchers believe that this strategy will ultimately allow biologists to monitor the activation of thousands of transcription factors, leading to a better understanding of the mechanisms underlying cell division and development. "Alternatively, since these nanosensors work directly in biological samples, we also believe that they could be used to screen and test new drugs that could, for example, inhibit transcription-factor binding activity responsible for the growth of tumor cells," said Plaxco.
This work was funded by the National Institute of Health, the Fond Québécois de la Recherche sur la Nature et les Technologies, the Italian Ministry of University and Research (MIUR) project "Futuro in Ricerca," and the Tri-County Blood Bank Santa Barbara Foundation.
† Top photo: A structure-switching nanosensor made from DNA (blue and purple) detects a specific transcription factor (green). Using these nanosensors, a team of researchers from UCSB has demonstrated the detection of transcription factors directly in cellular extracts. The researchers believe that their strategies will allow biologists to monitor the activity of thousands of transcription factors, leading to a better understanding of the mechanisms underlying cell division and development.
Credit: Peter Allen
Related Links