Scientists Get Glimpse of How the ‘Code' of Life May Have Emerged by Deciphering Molecular Puzzle
(Santa Barbara, Calif.) –– A portion of the "code" of life has been unraveled by a UC Santa Barbara graduate student from the town of Jojutla, Mexico.
Annia Rodriguez worked with John Perona, professor in UCSB's Department of Chemistry and Biochemistry, to decipher intramolecular communication within a large RNA-protein enzyme responsible for expressing the genetic code for the amino acid glutamine.
To their surprise, the experiments by Rodriguez captured a partial glimpse of how the genetic coding of life may have emerged. The results of the study are published in the journal Structure, published by CELL.
Life is based on the ability of all living cells to convert the genetic information in DNA, into the specific sequences of amino acids that make up the proteins that are the cell's workhorses. The key reaction in this decoding process is the attachment of a particular amino acid to one end of a small RNA molecule known as a transfer RNA. The enzyme that catalyzes this amino acid-RNA attachment is the aminoacyl-tRNA synthetase.
Rodriguez performed many laborious experiments in which she removed portions of the aminoacyl-tRNA synthetase that interact with the anticodon stem of the transfer RNA, far from the part of the enzyme that binds the amino acid. Using a biochemical approach known as rapid chemical quench kinetics, Rodriguez discovered that when she made these changes to the enzyme, the binding of the amino acid to the protein was strengthened, even though the amino acid binds far away from the positions where the changes were made.
"It is totally counterintuitive," said Perona. "Imagine if you had a car, and you took out a gear, and the car went faster. Why would you want that gear if it makes your car go slower?"
In all, Rodriguez found that separately removing seven different "gears" from a distant part of the molecule each caused the amino acid to bind more tightly to the aminoacyl-tRNA synthetase. Perona explained that this provides the first systematic analysis demonstrating long-range communication in an enzyme that depends on RNA for its function.
"So what we think is going on is that these enzyme-RNA interactions far from the amino acid binding site evolved together with the needs of the cell to respond to subtle cues from its environment – especially in terms of how much amino acid is available," said Perona. "It makes sense in terms of evolution."
Rodriguez is the first in her family to pursue a Ph.D., which she will complete this year. Now 28 years old, she began her career as a nurse in Cuernavaca, Mexico. Then she went on to obtain a B.S. in biochemical engineering at the Instituto Tecnológico de Zacatepec.
Graduation from her undergraduate program called for work at a research institution and she chose UCSB. Upon graduation, Rodriguez was offered a prestigious five-year scholarship with Mexico's Consejo Nacional de Ciencia y Technología (CONACYT) to continue her studies at UCSB.
Although her current research is not focused specifically on human health, Rodriguez said: "My interest in biochemistry started because I wanted to know the mechanisms by which drugs and medications worked inside the human body. I wanted to learn not just the signs and symptoms of disease, but how diseases are developed in a molecular level."
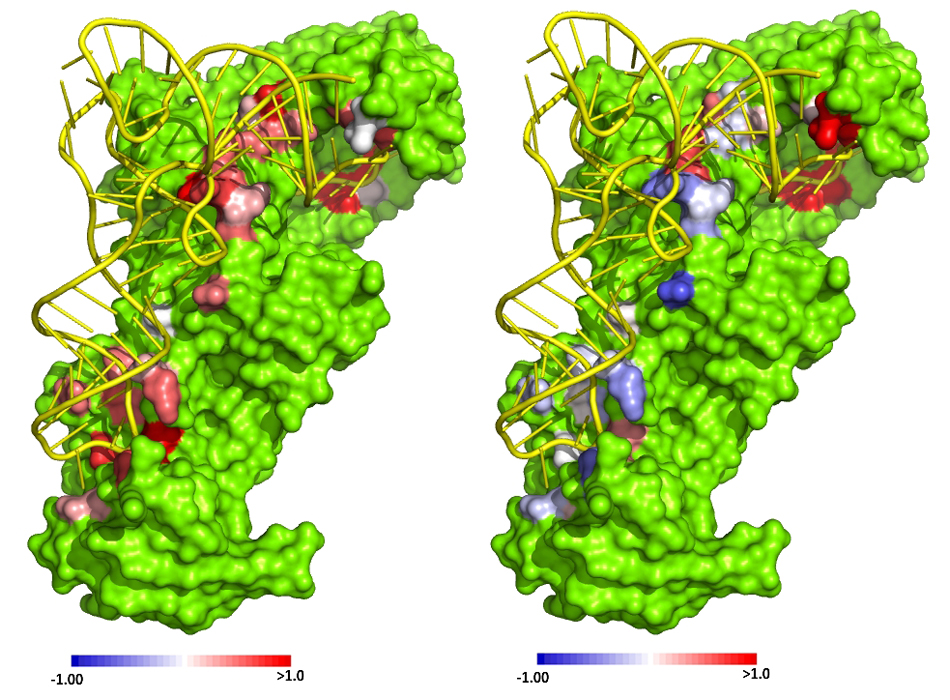
† Top photo: Crystal structure of glutaminyl tRNA synthetase (GlnRS, green) in complex with its substrate tRNA^Gln (yellow). Right panel: Color-coded residues depict favorable (blue) and unfavorable (red) effects on the free energy of glutamine binding from mutation at this position. Left panel: Effects of mutation on the ability of GlnRS to catalyze amino acid attachment to the tRNA. In this case all effects are unfavorable.
Credit: Annia Rodriguez/ John Perona / UCSB
†† Bottom photo: Annia Rodriguez and John Perona in front of the Rapid Quench Flow Machine used in the experiments.
Credit: George Foulsham, Office of Public Affairs, UCSB
Related Links